- Share
- Share on Facebook
- Share on X
- Share on LinkedIn
 |
#24-42. Sacchi, M., Sauter-Starace, F., Mailley, P., and Texier, I. (2024) Resorbable conductive materials for optimally interfacing medical devices with the living. Front Bioeng Biotechnol 12, 1294238, doi: 10.3389/fbioe.2024.1294238 |
 |
#24-41. Harrabi, R., Halbritter, T., Alarab, S., Chatterjee, S., Wolska-Pietkiewicz, M., Damodaran, K. K., van Tol, J., Lee, D., Paul, S., Hediger, S., Sigurdsson, S. T., Mentink-Vigier, F., and De Paepe, G. (2024) AsymPol-TEKs as efficient polarizing agents for MAS-DNP in glass matrices of non-aqueous solvents. Phys Chem Chem Phys 26, 5669-5682, doi: 10.1039/d3cp04271e |
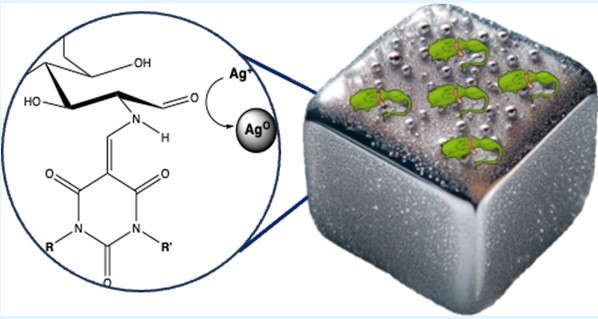 |
#24-40. Chen, B., Fragal, E. H., Faudry, E., and Halila, S. (2024) In Situ Growth of Silver Nanoparticles into Reducing-End Carbohydrate-Based Supramolecular Hydrogels for Antimicrobial Applications. ACS Appl. Mater. Interf. 16, 70818–70827, doi: 10.1021/acsami.4c17526 (text available in HAL) |
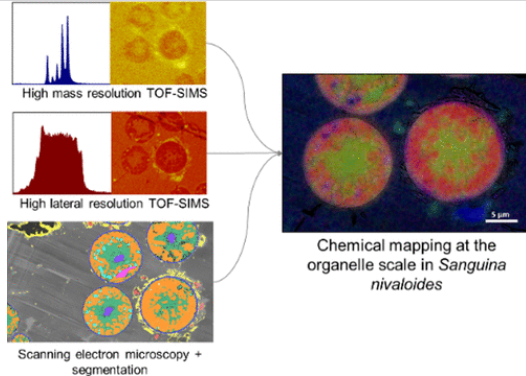 |
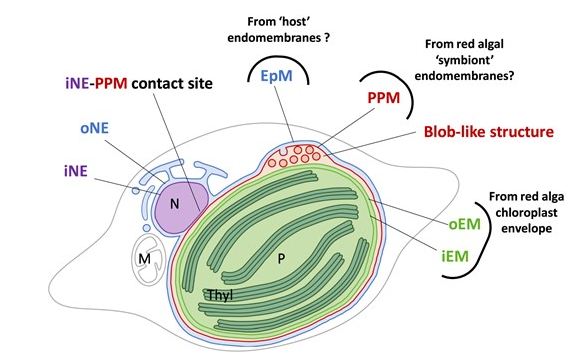
#24-39. Seydoux, C., Ezzedine, J. A., Larbi, G. S., Ravanel, S., Marechal, E., Barnes, J. P., and Jouneau, P. H. (2024) Subcellular ToF-SIMS Imaging of the Snow Alga Sanguina nivaloides by Combining High Mass and High Lateral Resolution Acquisitions. Anal. Chem., 96, 19917−19925, doi: 10.1021/acs.analchem.4c03826 |
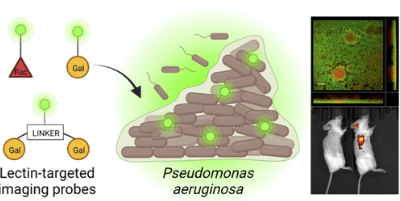 |
#24-38. Zahorska, E., Denig, L. M., Lienenklaus, S., Kuhaudomlarp, S., Tschernig, T., Lipp, P., Munder, A., Gillon, E., Minervini, S., Verkhova, V., Imberty, A., Wagner, S., and Titz, A. (2024) High-Affinity Lectin Ligands Enable the Detection of Pathogenic Pseudomonas aeruginosa Biofilms: Implications for Diagnostics and Therapy. JACS Au, 4, 4715–4728, doi: 10.1021/jacsau.4c00670 (text available in HAL) |
 |
#24-37. Gueguen, N., Seres, Y., Ciceron, F., Gros, V., Si Larbi, G., Falconet, D., Deragon, E., Gueye, S. D., Le Moigne, D., Schilling, M., Cussac, M., Petroutsos, D., Hu, H., Gong, Y., Michaud, M., Jouhet, J., Salvaing, J., Amato, A., and Marechal, E. (2024) Monogalactosyldiacylglycerol synthase isoforms play diverse roles inside and outside the diatom plastid. Plant Cell 36, 5023–5049, doi: 10.1093/plcell/koae275 (text available in HAL) |
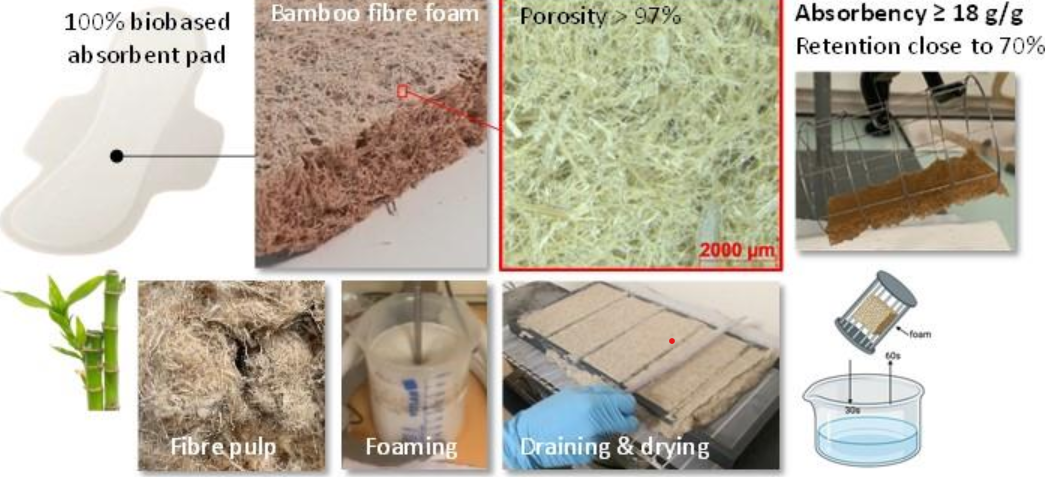 |
#24-36. Deville, M., Marietti, N., Viguié, J., Sillard, C., and Charlier, Q. (2024) Liquid Absorbent Bamboo Fiber Foams: Towards 100% Ligno-cellulosic Menstrual Absorbent Pads. BioResources 19, 9036-9048, doi: 10.15376/biores.19.4.9036-9048 |
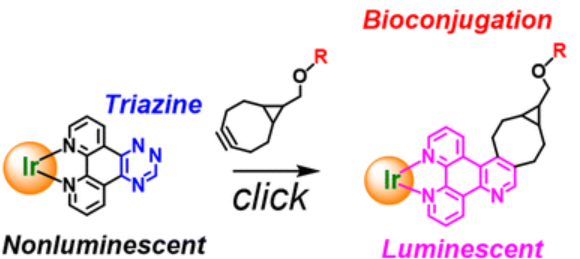 |
#24-35. Cooke, L., Gristwood, K., Adamson, K., Sims, M. T., Deary, M. E., Bruce, D., Antoniou, A. N., Walden, H. R., Knight, J. C., Antoine-Brunet, T., Joly, M., Goyard, D., Lanoe, P. H., Berthet, N., and Kozhevnikov, V. N. (2024) Annealing 1,2,4-triazine to iridium(III) complexes induces luminogenic behaviour in bioorthogonal reactions with strained alkynes. Dalton Trans 53, 15501-15508, doi: 10.1039/d4dt01499e (text available in HAL) |
 |
#24-34. Darmau, B., Hoang, A., Gross, A. J., and Texier, I. (2024) Water-based synthesis of dextran-methacrylate and its use to design hydrogels for biomedical applications. Eur. Polym. J. 221, 113515, doi: 10.1016/j.eurpolymj.2024.113515 |
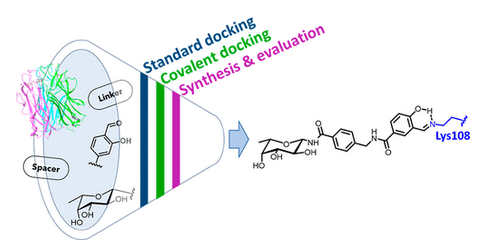 |
#24-33. Antonini, G., Bernardi, A., Gillon, E., Dal Corso, A., Civera, M., Belvisi, L., Varrot, A., and Mazzotta, S. (2024) Achieving High Affinity for a Bacterial Lectin with Reversible Covalent Ligands. J. Med. Chem., 67, 19546−19560, doi: 10.1021/acs.jmedchem.4c01876 (text available in HAL) |
| #24-32. Kim, M., Jorge, G. L., Aschern, M., Cuine, S., Bertrand, M., Mekhalfi, M., Putaux, J. L., Yang, J. S., Thelen, J. J., Beisson, F., Peltier, G., and Li-Beisson, Y. (2024) The DYRKP1 kinase regulates cell wall degradation in Chlamydomonas by inducing matrix metalloproteinase expression. Plant Cell, 36, 4988–5003, doi: 10.1093/plcell/koae271(text available in HAL) | |
| #24-31. Whitehead, A. K., Wang, Z., Boustany, R. J., Vives, R. R., Lazartigues, E., Liu, J., Siggins, R. W., and Yue, X. (2024) Myeloid Deficiency of Heparan Sulfate 6-O-Endosulfatases Impairs Bone Marrow Hematopoiesis. Matrix Biol.,134, 107-118 doi: 10.1016/j.matbio.2024.10.002 | |
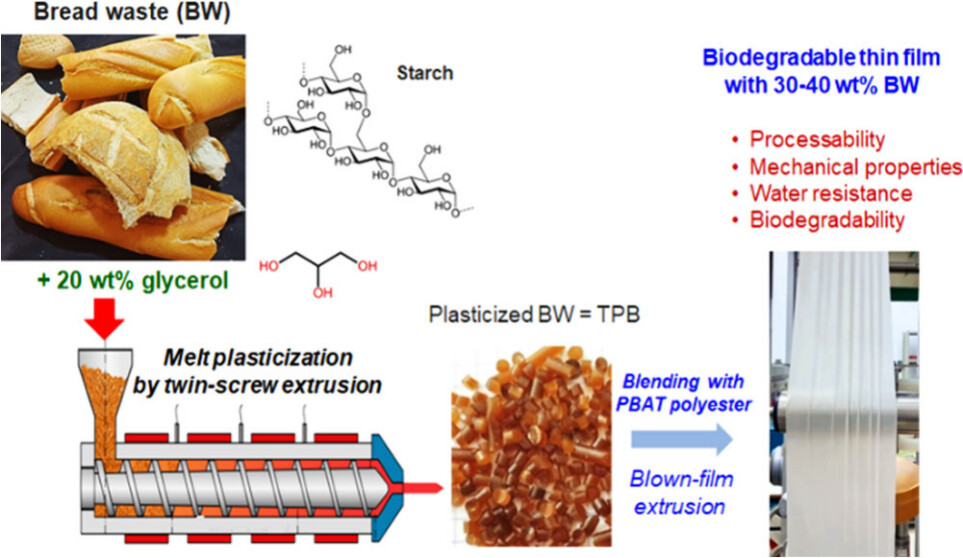 |
#24-30. Bouzidi, S., Lévèque, J.-M., Magnin, A., Molina-Boisseau, S., Putaux, J.-L., and Boufi, S. (2024) Thermoplasticized Bread Waste and Poly(butylene adipate-co-terephthalate) Blends: A Sustainable Alternative to Starch in Eco-Friendly Packaging. ACS Sustainable Chem. Eng.,12,15268 - 15281, doi: 10.1021/ma00077a003 (text available in HAL) |
| #24-29. Leite, L., Le Gars, M., Azeredo, H., Moreira, F., Mattoso, L., and Bras, J. (2024) Insights into the effect of carboxylated cellulose nanocrystals on mechanical and barrier properties of gelatin films for flexible packaging applications. Int. J. Biol. Macromol. 250, 135726, doi: 10.1016/j.ijbiomac.2024.135726 | |
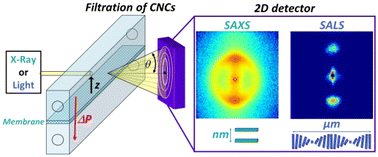 |
#24-28. Mandin, S., Metilli, L., Karrouch, M., Bleses, D., Lancelon-Pin, C., Sailler, P., Chevremont, W., Paineau, E., Putaux, J. L., Hengl, N., Jean, B., and Pignon, F. (2024) Multiscale study of the chiral self-assembly of cellulose nanocrystals during the frontal ultrafiltration process. Nanoscale, 16, 19100–19115, doi: 10.1039/d4nr02840f (text available in HAL) |
| #24-27. Saulais, M., Salem, S., Sillard, C., Choisy, P., and Dufresne, A. (2024) Green synthesis of sacrificial UV-sensitive core and biobased shell for obtaining optically hollow nanoparticles. J. Colloid Interface Sci. 678, 971-983, doi: 10.1016/j.jcis.2024.08.260 | |
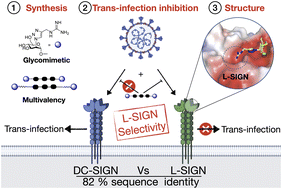 |
#24-26. Delaunay, C., Pollastri, S., Thepaut, M., Cavazzoli, G., Belvisi, L., Bouchikri, C., Labiod, N., Lasala, F., Gimeno, A., Franconetti, A., Jimenez-Barbero, J., Arda, A., Delgado, R., Bernardi, A., and Fieschi, F. (2024) Unprecedented selectivity for homologous lectin targets: differential targeting of the viral receptors L-SIGN and DC-SIGN. Chem. Sci. 15, 15352-66, doi: 10.1039/d4sc02980a (text available in HAL) |
| #24-25. Freville, E., Zeno, E., Meyer, V., Carré, B., Terrien, M., Mauret, E., and Bras, J. (2024) Process stability optimization of the twin‑screw extrusion adapted for concentrated cellulose fibrillation. J. Mater. Sci. 59, 15904–15919, doi: 10.1007/s10853-024-10115-7 | |
| #24-24. Le Pennec, J. L., Guibert, A., Gurram, R., Delon, A., Vives, R. R., and Migliorini, E. (2024) BMP2 Binds Non-Specifically to PEG-Passivated Biomaterials and Induces pSMAD 1/5/9 Signalling. Macromol Biosci, e2400169, doi: 10.1002/mabi.202400169 (text available in HAL) | |
| #24-23. Soullard, L., Schlepp, A., Buret, R., Lancelon‑Pin, C., Nonglaton, G., Texier, I., Jean, B., and Rolere, S. (in press) Towards the 3D printing of innovative hydrogel scaffolds through vat polymerization techniques using methacrylated carboxymethylcellulose aqueous formulations. Progress in Additive Manufacturing, doi: 10.1007/s40964-024-00744-4 (text available in HAL) | |
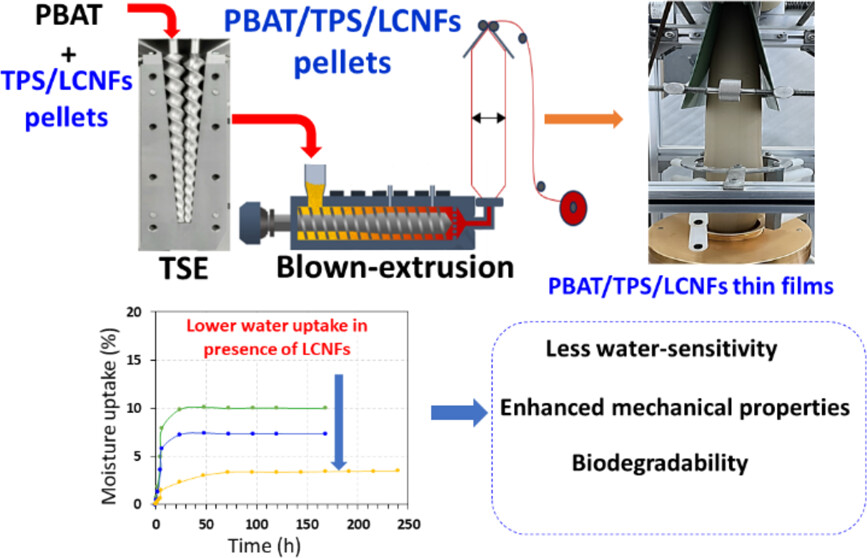 |
#24-22. Aouay, M., Magnin, A., Lancelon-Pin, C., Putaux, J.-L., and Boufi, S. (2024) Mitigating the Water Sensitivity of PBAT/TPS Blends through the Incorporation of Lignin-Containing Cellulose Nanofibrils for Application in Biodegradable Films. ACS Sustainable Chem. Eng. 12, 10805−10819, doi: 10.1021/acssuschemeng.4c02245 (text available in HAL) |
| #24-21. Freville, E., Sergienko, J. P., Mujica, R., Rey, C., and Bras, J. (2024) Novel technologies for producing tridimensional cellulosic materials for packaging: A review. Carbohydr. Polym. 342, 122413, doi: 10.1016/j.carbpol.2024.122413 (text available in HAL) | |
| #24-20. van Trijp, J. P., Hribernik, N., Lim, J. H., Dal Colle, M. C. S., Vazquez Mena, Y., Ogawa, Y., and Delbianco, M. (2024) Enzyme-triggered assembly of glycan nanomaterials. Angew. Chem. Int. Ed. Engl. 63,, e202410634, doi: 10.1002/anie.202410634 | |
| #24-19. Bouzidi, Y., Bosco, M., Gao, H., Pradeau, S., Matheron, L., Chantret, I., Busca, P., Fort, S., Gravier-Pelletier, C., and Moore, S. E. H. (2024) Transport of N-acetylchitooligosaccharides and fluorescent N-acetylchitooligosaccharide analogs into rat liver lysosomes. Glycobiology 34, cwad099, doi: 10.1093/glycob/cwad099 (text available in HAL) | |
| #24-18. Doineau, E., Pucci, M. F., Cathala, B., Benezet, J.-C., Bras, J., and Le Moigne, N. (2024) Multiscale analysis of hierarchical flax-epoxy biocomposites with nanostructured interphase by xyloglucan and cellulose nanocrystals. Composites Part A 184, 108270, doi: 10.1016/j.compositesa.2024.108270 (text available in HAL) | |
| #24-17. Zhu, J., Li, S., Chen, W., Xu, X., Wang, X., Wang, X., Han, J., Jouhet, J., Amato, A., Marechal, E., Hu, H., Allen, A. E., Gong, Y., and Jiang, H. (2024) Delta-5 elongase knockout reduces docosahexaenoic acid and lipid synthesis and increases heat sensitivity in a diatom. Plant Physiol., 191, 1356–1373, doi: 10.1093/plphys/kiae297 (text available in HAL) | |
| #24-16. Abdallah, A., Gillon, E., Rannou, P., Imberty, A., and Halila, S. (2024) Microwave-Assisted Synthesis of β-N-Aryl Glycoamphiphiles with Diverse Supramolecular Assemblies and Lectin Accessibility. Bioconjug. Chem. 35,1200-1206 doi: 10.1021/acs.bioconjchem.4c00224 (text available in HAL) | |
| #24-15. Ribeiro, D. O., Bonnardel, F., Palma, A. S., Carvalho, A. L. M., and Perez, S. (2024) CBMcarb-DB: interface of the three-dimensional landscape of carbohydrate-binding modules. Carbohydr. Chem. 46, 1-22, doi: 10.1039/BK9781837672844-00001 | |
| #24-14. Fan, Y., El Rhaz, A., Maisonneuve, S., Gillon, E., Fatthalla, M., Le Bideau, F., Laurent, G., Messaoudi, S., Imberty, A., and Xie, J. (2024) Photoswitchable glycoligands targeting Pseudomonas aeruginosa LecA. Beilstein J. Org. Chem. 20, 1486–1496, doi: 10.3762/bxiv.2024.15 (text available in HAL) | |
| #24-13. Le Pennec, J., Makshakova, O., Nevola, P., Fouladkar, F., Gout, E., Machillot, P., Friedel-Arboleas, M., Picart, C., Pérez, S., Vortkamp, A., Vivès, R. R., and Migliorini, E. (2024) Glycosaminoglycans exhibit distinct interactions and signaling with BMP2 according to their nature and localization. Carbohydr. Polym. 341, 122294, doi: 10.1016/j.carbpol.2024.122294 (text available in HAL) | |
| #24-12. Zhang, J., Sun, J., Chiu, C. H., Landry, D., Li, K., Wen, J., Mysore, K. S., Fort, S., Lefebvre, B., Oldroyd, G. E. D., and Feng, F. (2024) A receptor required for chitin perception facilitates arbuscular mycorrhizal associations and distinguishes root symbiosis from immunity. Curr. Biol. 8, 1705-1717, doi: 10.1016/j.cub.2024.03.015 (text available in HAL) | |
| #24-11. Mandin, S., Metilli, L., Karrouch, M., Lancelon-Pin, C., Putaux, J.-L., Chevremont, W., Paineau, E., Hengl, N., Jean, B., and Pignon, F. (2024) Chiral nematic nanocomposites with pitch gradient elaborated by filtration and ultraviolet curing of cellulose nanocrystal suspensions. Carbohydr. Polym. 337, 122162, doi: 10.1016/j.carbpol.2024.122162 (text available in HAL) | |
|
#24-10. Le Pennec, J., Guibert, A., Vivès, R. R., and Migliorini, E. (Preprint) BMP2 binds non-specifically to PEG-passivated biomaterials and induces substantial signaling. BioRXiv, doi: 10.1101/2024.03.14.585026
|
|
| #24-09. Nieto-Fabregat, F., Zhu, Q., Vivès, C., Zhang, Y., Marseglia, A., Chiodo, F., Thépaut, M., Rai, D., Kulkarni, S. S., Di Lorenzo, F., Molinaro, A., Marchetti, R., Fieschi, F., Xiao, G., Yu, B., and Silipo, A. (2024) Atomic-Level Dissection of DC-SIGN Recognition of Bacteroides vulgatus LPS Epitopes. JACS Au 4, 697–712, doi: 10.1021/jacsau.3c00748 (text available in HAL) | |
| #24-08. Herrera-Gonzalez, I., Gonzalez-Cuesta, M., Thepaut, M., Laigre, E., Goyard, D., Rojo, J., Garcia Fernandez, J. M., Fieschi, F., Renaudet, O., Nieto, P. M., and Ortiz Mellet, C. (2024) High Mannose Oligosaccharide Hemimimetics that Recapitulate the Conformation and Binding Mode to Concanavalin A, DC-SIGN and Langerin. Chemistry, 30, e202303041, doi: 10.1002/chem.202303041 (text available in HAL) | |
| #24-07. Pennec, J. L., Picart, C., Vives, R. R., and Migliorini, E. (2024) Sweet but Challenging: Tackling the Complexity of GAGs with Engineered Tailor-Made Biomaterials. Adv Mater, 36, e2312154, doi: 10.1002/adma.202312154 (text available in HAL) | |
| #24-06. Nieto-Fabregat, F., Marseglia, A., Thepaut, M., Kleman, J. P., Abbas, M., Le Roy, A., Ebel, C., Maalej, M., Simorre, J. P., Laguri, C., Molinaro, A., Silipo, A., Fieschi, F., and Marchetti, R. (2024) Molecular recognition of Escherichia coli R1-type core lipooligosaccharide by DC-SIGN. iScience 27, 108792, doi: 10.1016/j.isci.2024.108792 (text available in HAL) | |
| #24-05. Pignon, F., Guilbert, E., Mandin, S., Hengl, N., Karrouch, M., Jean, B., Putaux, J. L., Gibaud, T., Manneville, S., and Narayanan, T. (2024) Orthotropic organization of a cellulose nanocrystal suspension realized via the combined action of frontal ultrafiltration and ultrasound as revealed by in situ SAXS. J. Colloid Interface Sci. 659, 914-925, doi: 10.1016/j.jcis.2023.12.164 (text available in HAL) | |
| #24-04. You, L., Polonska, A., Jasieniecka-Gazarkiewicz, K., Richard, F., Jouhet, J., Marechal, E., Banas, A., Hu, H., Pan, Y., Hao, X., Jin, H., Allen, A. E., Amato, A., and Gong, Y. (2024) Two plastidial lysophosphatidic acid acyltransferases differentially mediate the biosynthesis of membrane lipids and triacylglycerols in Phaeodactylum tricornutum. New Phytol., 241, 1543-1558, doi: 10.1111/nph.19434 (text available in HAL) | |
| #24-03. Huang, T., Pan, Y., Marechal, E., and Hu, H. (2024) Proteomes reveal the lipid metabolic network in the complex plastid of Phaeodactylum tricornutum. Plant J., 117, 385-403, doi: 10.1111/tpj.16477 (text available in HAL) | |
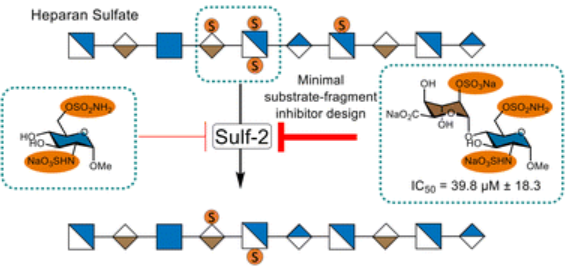 |
#24-02. Kennett, A., Epple, S., van der Valk, G., Georgiou, I., Gout, E., Vives, R. R., and Russell, A. (2024) Modified minimal-size fragments of Heparan Sulfate as inhibitors of endosulfatase-2 (Sulf-2). ChemComm, 60, 436, doi: 10.1039/D3CC02565A (text available in HAL) |
| #24-01. Schnider, B., M'Rad, Y., El Ahmadie, J., de Brevern, A., Imberty, A., and Lisacek, F. (2024) HumanLectome, an update of UniLectin for the annotation and prediction of human lectins. Nucleic Acids Res., 52, D1683–D1693, doi: 10.1093/nar/gkad905 (text available in HAL) |
- Share
- Share on Facebook
- Share on X
- Share on LinkedIn