 |
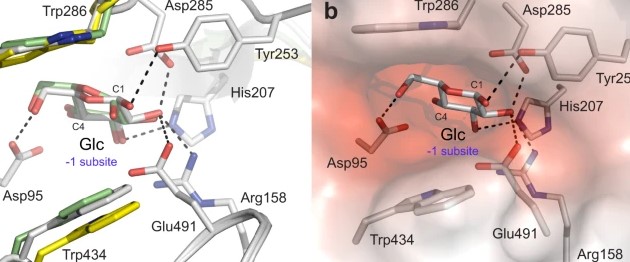
#22-55. Luang, S., Fernández-Luengo, X., Nin-Hill, A., Streltsov, V. A., Schwerdt, J. G., Alonso-Gil, S., Cairns, J. R. K., Pradeau, S., Fort, S., Maréchal, J.-D., Masgrau, L., Rovira, C., and Hrmova, M. (2022) The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases. Nat. Commun. 13, 5577, doi: 10.1038/s41467-022-33180-5 (full text available in HAL) |
 |
#22-54. Villalobos Solis, M. I., Engle, N. L., Spangler, M. K., Cottaz, S., Fort, S., Maeda, J., Ané, J.-M., Tschaplinski, T. J., Labbé, J. L., Hettich, R. L., Abraham, P. E., and Rush, T. A. (2022) Expanding the Biological Role of Lipo-Chitooligosaccharides and Chitooligosaccharides in Laccaria bicolor Growth and Development. Front. Fungal Biol. 3, 808758, doi: 10.3389/ffunb.2022.808578 (full text available in HAL) |
 |
#22-53.Goyard, D., Ortiz, A. M., Boturyn, D., and Renaudet, O. (2022) Multivalent glycocyclopeptides: conjugation methods and biological applications. Chem. Soc. Rev. 51, 8756-8783, doi: 10.1039/d2cs00640e (full text available in HAL) |
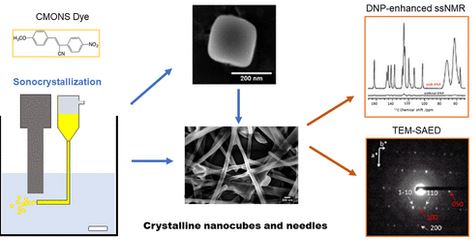 |
#22-52. Cattoën, X., Kumar, A., Dubois, F., Vaillant, C., Matta-Seclén, M., Leynaud, O., Kodjikian, S., Hediger, S., Paëpe, G. D., and Ibanez, A. (2022) Sonocrystallization of CMONS needles and nanocubes: mechanistic studies and advanced crystallinity characterization by combining X‑ray and electron diffractions with DNP-enhanced NMR. Cryst. Growth Des. 22, 2181−2191, doi: https://doi.org/10.1021/acs.cgd.1c01246 |
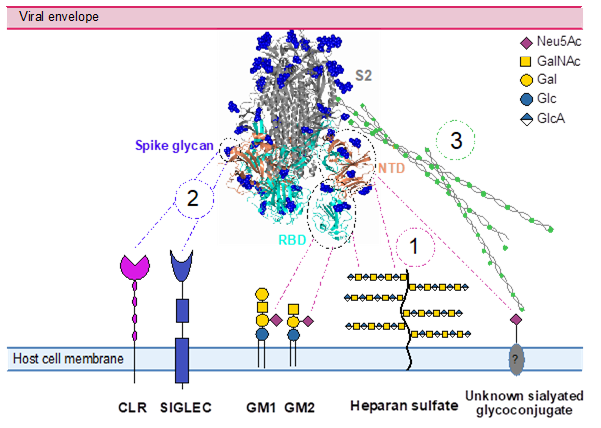 |
#22-51. Kuhaudomlarp, S., and Imberty, A. (2022) Involvement of sialoglycans in SARS-COV-2 infection: opportunities and challenges for glyco-based inhibitors. IUBMB Life 17, 1253-1263, doi: 10.1002/iub.2692 (full text available in HAL) |
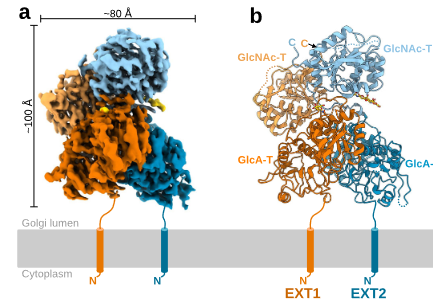 |
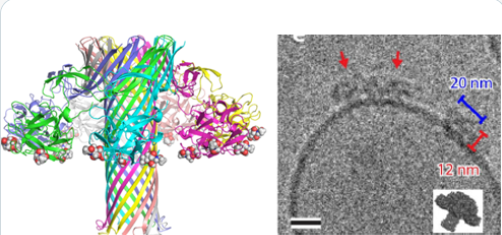
#22-50. Leisico, F., Omeiri, J., Le Narvor, C., Beaudouin, J., Hons, M., Fenel, D., Schoehn, G., Coute, Y., Bonnaffe, D., Sadir, R., Lortat-Jacob, H., and Wild, R. (2022) Structure of the human heparan sulfate polymerase complex EXT1-EXT2. Nat. Commun. 13, 7110, doi: 10.1038/s41467-022-34882-6 |
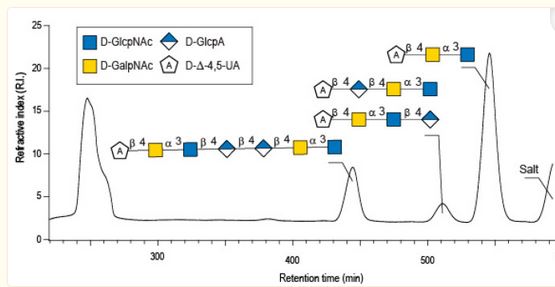 |
#22-49. Drouillard, S., Poulet, L., Boisset, C., Delbarre-Ladrat, C., and Helbert, W. (2022) NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629. Mar. Drugs 20, 731, doi: 10.3390/md20120731 (full text available in HAL) |
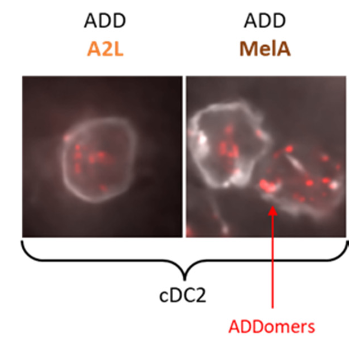 |
#22-48. Besson, S., Laurin, D., Chauviere, C., Thepaut, M., Kleman, J. P., Pezet, M., Manches, O., Fieschi, F., Aspord, C., and Fender, P. (2022) Adenovirus-inspired virus-like-particles displaying melanoma tumor antigen specifically target human DC subsets and trigger antigen-specific immune responses. Biomedicines 10, doi: 10.3390/biomedicines10112881 |
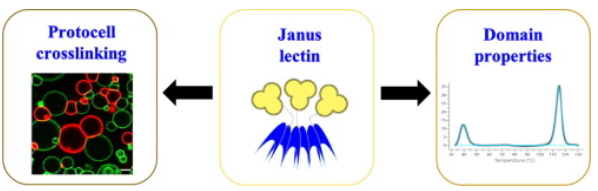 |
#22-47. Notova, S., Siukstaite, L., Rosato, F., Vena, F., Audfray, A., Bovin, N., Landemarre, L., Römer, W., and Imberty, A. (2022) Extending Janus lectins architecture: characterization and application to protocells. Comput. Struct. Biotechnol. J. 20, 6108–6119, doi: 10.1016/j.csbj.2022.11.005 (full text available in HAL) |
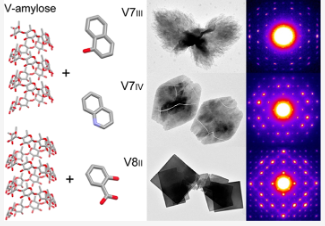 |
#22-46. Le, C. A. K., Ogawa, Y., Grimaud, F., Nishiyama, Y., Morfin, I., Potocki-Veronese, G., Choisnard, L., Wouessidjewe, D., and Putaux, J.-L. (2022) Helical Inclusion Complexes of Amylose with Aromatic Compounds: Crystallographic Evidence for New V‑Type Allomorphs. Cryst. Growth Design 22, 7079-7089, doi: 10.1021/acs.cgd.2c00771 (full text available in HAL) |
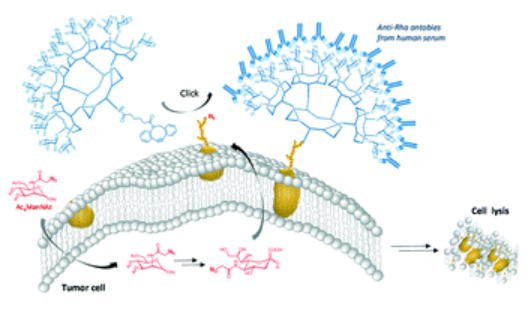 |
#22-45. Goyard, D., Diriwari, P. I., and Berthet, N. (2022) Metabolic labelling of cancer cells with glycodendrimers stimulate immune-mediated cytotoxicity. RSC Med Chem 13, 72-78, doi: 10.1039/d1md00262g (full text available in HAL) |
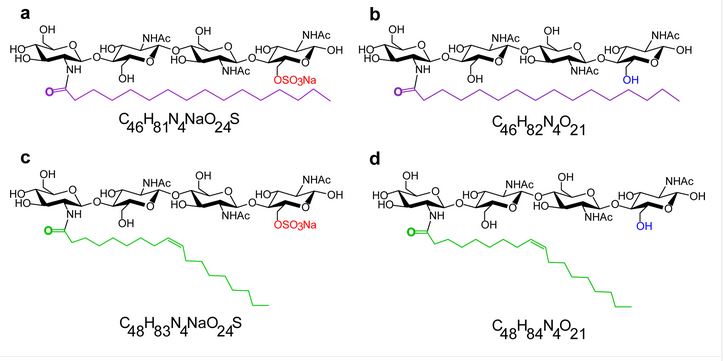 |
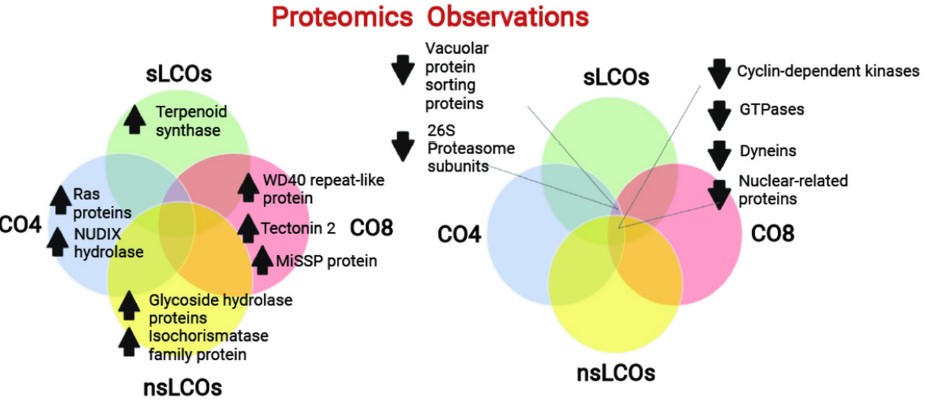
#22-44. Rush, T. A., Tannous, J., Lane, M. J., Gopalakrishnan Meena, M., Carrell, A. A., Golan, J. J., Drott, M. T., Cottaz, S., Fort, S., Ane, J. M., Keller, N. P., Pelletier, D. A., Jacobson, D. A., Kainer, D., Abraham, P. E., Giannone, R. J., and Labbe, J. L. (2022) Lipo-Chitooligosaccharides Induce Specialized Fungal Metabolite Profiles That Modulate Bacterial Growth. mSystems 7, e0105222, doi: 10.1128/msystems.01052-22 |
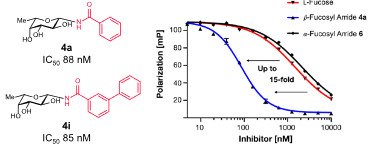 |
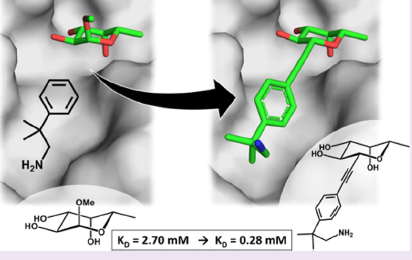
#22-43. Mala, P., Siebs, E., Meiers, J., Rox, K., Varrot, A., Imberty, A., and Titz, A. (2022) Discovery of N-beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB. J. Med. Chem. 65, 14180-14200, doi: 10.1021/acs.jmedchem.2c01373 (full text available in HAL) |
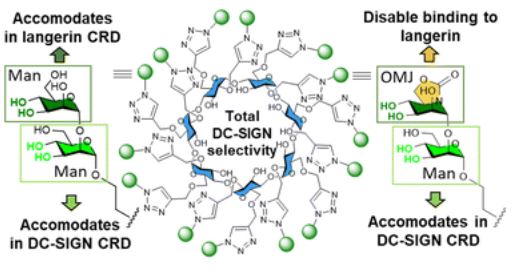 |
#22-42. Herrera-Gonzalez, I., Thepaut, M., Sanchez-Fernandez, E. M., di Maio, A., Vives, C., Rojo, J., Garcia Fernandez, J. M., Fieschi, F., Nieto, P. M., and Ortiz Mellet, C. (2022) Mannobioside biomimetics that trigger DC-SIGN binding selectivity. Chem Commun 58, 12086-12089, doi: 10.1039/d2cc04478a (full text available in HAL) |
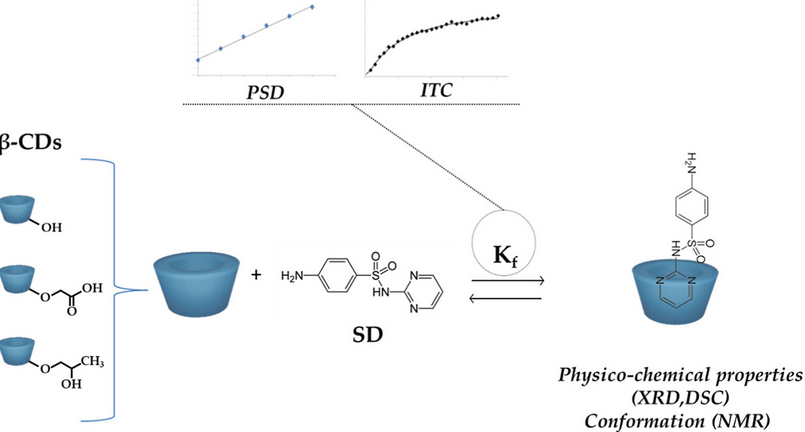 |
#22-41. Michel, B., Heggset, E. B., Syverud, K., Dufresne, A., and Bras, J. (2022) Inclusion complex formation between sulfadiazine and various modified β-cyclodextrins and characterization of the complexes. J. Drug Deliv. Sci.Technol. 76, 103814, doi: 10.1016/j.jddst.2022.103814 |
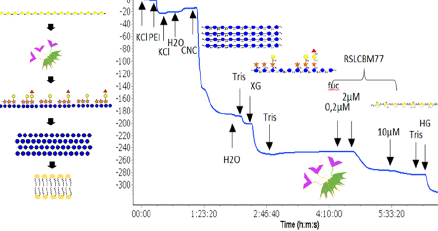 |
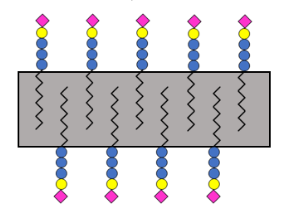
#22-40. Notova, S., Cannac, N., Rabagliati, L., Touzard, M., Mante, J., Navon, Y., Coche-Guerente, L., Lerouxel, O., Heux, L., and Imberty, A. (2022) Building an artificial plant cell wall on a lipid bilayer by assembling polysaccharides and engineered proteins. ACS Synthetic Biology 11, 3516-3528 doi: 10.1021/acssynbio.2c00404 (full text available in HAL) |
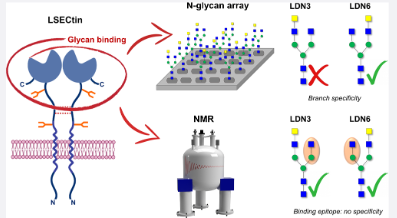 |
#22-39. Bertuzzi, S., Peccati, F., Serna, S., Artschwager, R., Notova, S., Thépaut, M., Jiménez-Osés, G., Fieschi, F., Reichardt, N. C., Jiménez-Barbero, J., and Ardá, A. (2022) Immobilization of biantennary N‑glycans leads to branch specific epitope recognition by LSECtin. ACS Central Science 8, 1415-1423, doi: 10.1021/acscentsci.2c00719 (full text available in HAL) |
 |
#22-38. Bermeo, R., Lal, K., Ruggeri, D., Lanaro, D., Mazzotta, S., Vasile, F., Imberty, A., Belvisi, L., Varrot, A., and Bernardi, A. (2022) Targeting a Multidrug-Resistant Pathogen: First Generation Antagonists of Burkholderia cenocepacia's BC2L-C Lectin. ACS Chem. Biol. 17, 2899-2910 doi: 10.1021/acschembio.2c00532 (full text available in HAL) |
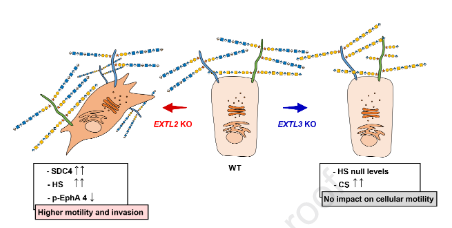 |
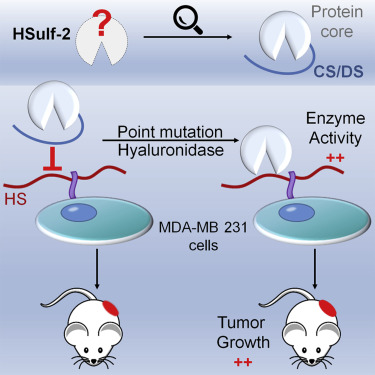
#22-37. Marques, C., Pocas, J., Gomes, C., Faria-Ramos, I., Reis, C. A., Vives, R. R., and Magalhaes, A. (2022) Glycosyltransferases EXTL2 and EXTL3 cellular balance dictates Heparan Sulfate biosynthesis and shapes gastric cancer cell motility and invasion. J. Biol. Chem., 102546, doi: 10.1016/j.jbc.2022.102546 (full text available in HAL) |
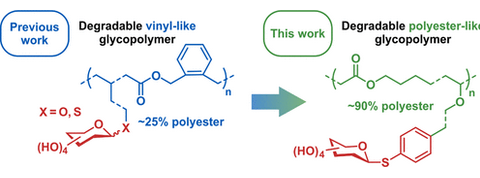 |
#22-36. Pesenti, T., Domingo-Lopez, D., Gillon, E., Ibrahim, N., Messaoudi, S., Imberty, A., and Nicolas, J. (2022) Degradable glycopolyester-like nanoparticles by radical ring-opening polymerization. Biomacromolecules 23, 4015–4028, doi: 10.1021/acs.biomac.2c00851 (full text available in HAL) |
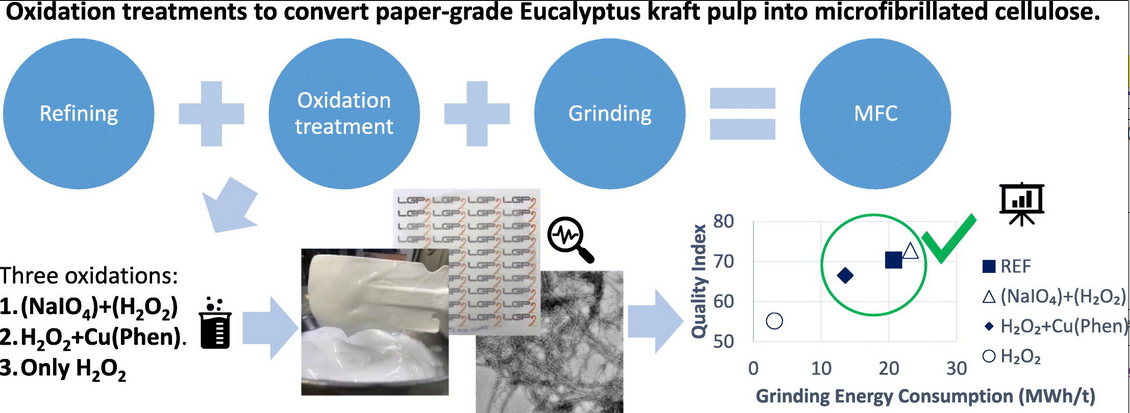 |
#22-35. Vera-Loor, A., Rigou, P., Marlin, N., Mortha, G., and Dufresne, A. (2022) Oxidation treatments to convert paper-grade Eucalyptus kraft pulp into microfibrillated cellulose. Carbohydr. Polym. 296, 119946, doi: 10.1016/j.carbpol.2022.119946 |
 |
#22-34. Notova, S., Bonnardel, F., Rosato, F., Siukstaite, L., Schwaiger, J., Bovin, N., Varrot, A., Römer, W., Lisacek, F., and Imberty, A. (2022) The choanoflagellate pore-forming lectin SaroL-1 punches holes in cancer cells by targeting tumor-related glycosphingolipid Gb3. Communications Biology 5, 594, doi: 10.1038/s42003-022-03869-w (full text available in HAL) |
 |
#22-33. Richard, E., Traversier, A., Julien, T., Rosa-Calatrava, M., Putaux, J. L., Jeacomine, I., and Samain, E. (2022) Biotechnological production of sialylated solid lipid microparticles as inhibitors of influenza A virus infection. Glycobiology 32, 949-961, doi: 10.1093/glycob/cwac054 (full text available in HAL) |
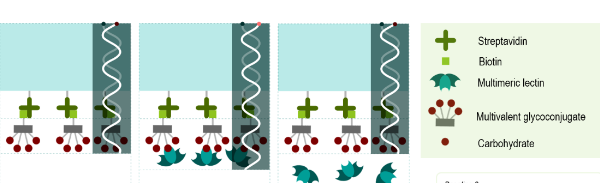 |
#22-32. Picault, L., Laigre, E., Gillon, E., Tiertant, C., Renaudet, O., Imberty, A., Goyard, D., and Dejeu, J. (2022) Characterization of the interaction of multivalent glycosylated ligands with bacterial lectins by BioLayer interferometry. Glycobiology 32, 886-896, doi: 10.1093/glycob/cwac047 (full text available in HAL) |
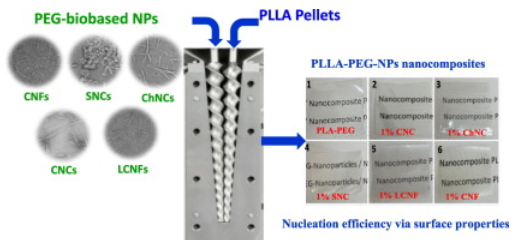 |
#22-31. Aouay, M., Magnin, A., Putaux, J. L., and Boufi, S. (2022) Biobased nucleation agents for poly-L-(lactic acid) - Effect on crystallization, rheological and mechanical properties. Int. J. Biol. Macromol. 219, 588-600, doi: 10.1016/j.ijbiomac.2022.07.069 (full text available in HAL) |
 |
#22-30. Perez, S., and Makshakova, O. (2022) Multifaceted Computational Modeling in Glycoscience. Chem. Rev. 122, 15914-15970, doi: 10.1021/acs.chemrev.2c00060 (full text available in HAL) |
 |
#22-29. Denamur, S., Chazeirat, T., Maszota-Zieleniak, M., Vivès, R. R., Saidi, A., Zhang, F., Linhardt, R. J., Labarthe, F., Samsonov, S. A., Lalmanach, G., and Lecaille, F. (2022) Binding of heparan sulfate to human cystatin C modulates inhibition of cathepsin L: Putative consequences in mucopolysaccharidosis. Carbohydr. Polym. 293, 119734, doi: 10.1016/j.carbpol.2022.119734 (full text available in HAL) |
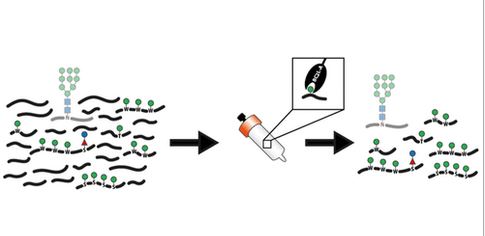 |
#22-28. Huette, H. J., Tiemann, B., Shcherbakova, A., Grote, V., Hoffmann, M., Povolo, L., Lommel, M., Strahl, S., Vakhrushev, S. Y., Rapp, E., Buettner, F. F. R., Halim, A., Imberty, A., and Bakker, H. (2022) A Bacterial Mannose Binding Lectin as a Tool for the Enrichment of C- and O-Mannosylated Peptides. Anal. Chem. 94, 7329-7338, doi: 10.1021/acs.analchem.2c00742 (full text available in HAL) |
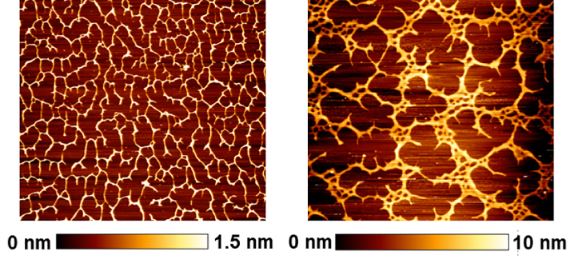 |
#22-27. Zykwinska, A., Makshakova, O., Gélébart, P., Sinquin, C., Stephant, N., Colliec-Jouault, S., Perez, S., and Cuenot, S. (2022) Interactions between infernan and calcium: From the molecular level to the mechanical properties of microgels. Carbohydr. Polym. 292, 119629, doi: 10.1016/j.carbpol.2022.119629 (full text available in HAL) |
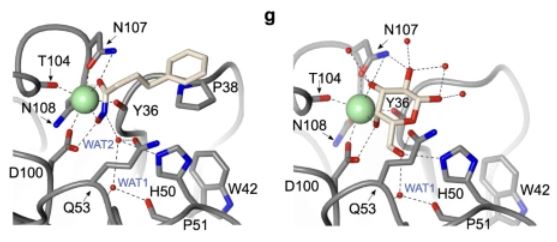 |
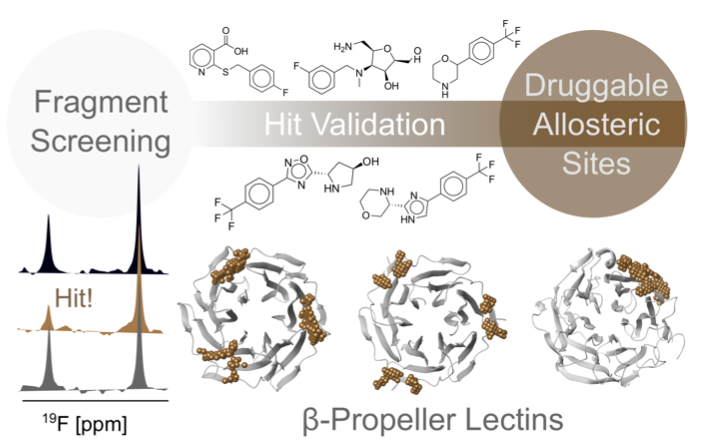
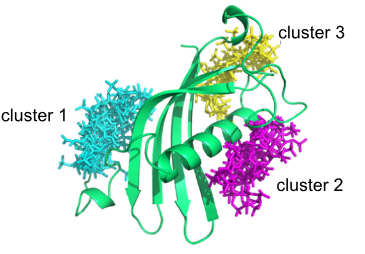
#22-26. Shanina, E., Kuhaudomlarp, S., Siebs, E., Fuchsberger, F., Denis, M., da Silva Figueiredo Celestino Gomes, P., Clausen, M. H., Seeberger, P. H., Rognan, D., Titz, A., Imberty, A., and Rademacher, C. (2022) Targeting undruggable carbohydrate recognition sites through focused fragment library design. Commun. Chem. 5, 64, doi: 10.1038/s42004-022-00679-3 (full text available in HAL) |
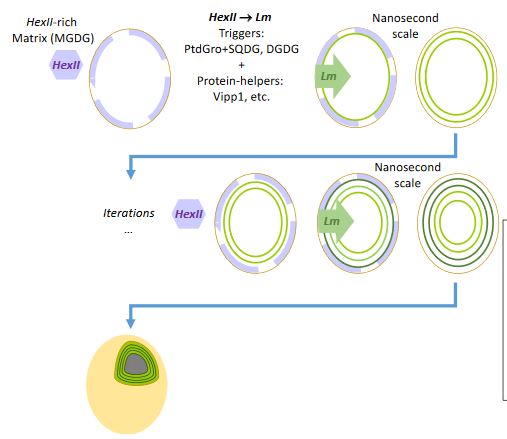 |
#22-25. Guéguen, N., and Maréchal, E. (2022) Origin of Cyanobacteria thylakoids via a non-vesicular glycolipid phase transition and impact on the Great Oxygenation Event J. Exp. Bot. 73, 2721–2734, doi: 10.1093/jxb/erab429 (full text available in HAL) |
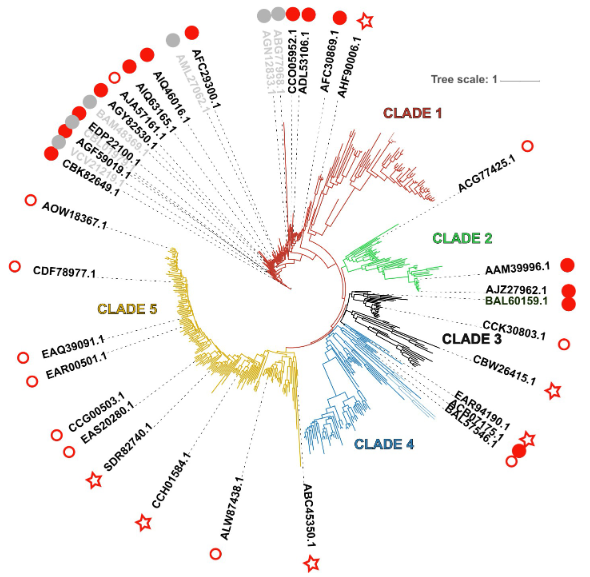 |
#22-24. Couturier, M., Touvrey-Loiodice, M., Terrapon, N., Drula, E., Buon, L., Chirat, C., Henrissat, B., and Helbert, W. (2022) Functional exploration of the glycoside hydrolase family GH113. PLoS One 17, e0267509, doi: 10.1371/journal.pone.0267509 (full text available in HAL) |
 |
#22-23. Chow, W. Y., De Paepe, G., and Hediger, S. (2022) Biomolecular and Biological Applications of Solid-State NMR with Dynamic Nuclear Polarization Enhancement. Chem. Rev. 122, 9795–9847, doi: 10.1021/acs.chemrev.1c01043 |